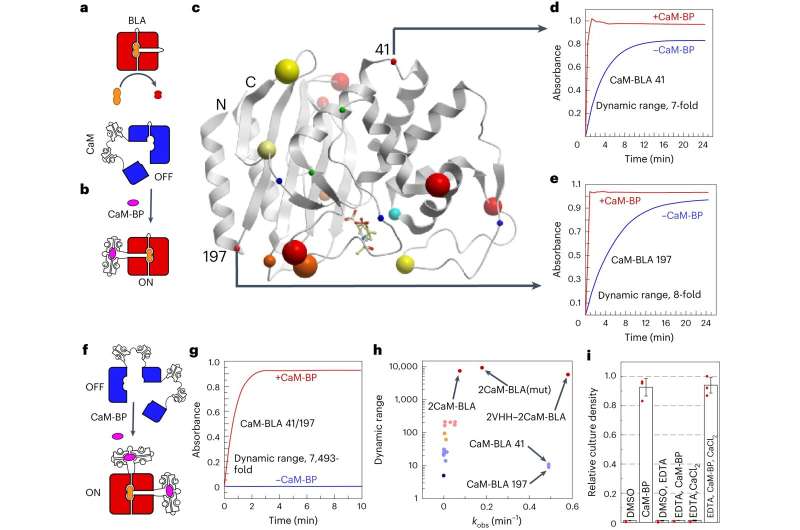
QUT researchers have developed a new approach for designing molecular ON-OFF switches based on proteins which can be used in a multitude of biotechnological, biomedical and bioengineering applications.
The research team demonstrated that this novel approach allows them to design and build faster and more accurate diagnostic tests for detecting diseases, monitoring water quality and detecting environmental pollutants.
Professor Kirill Alexandrov, of the QUT School of Biology and Environmental Science, lead scientist on the CSIRO-QUT Synthetic Biology Alliance and a researcher with the ARC Centre of Excellence in Synthetic Biology, said that the new technique published in Nature Nanotechnology demonstrated that protein switches could be engineered in a predictable way.
Professor Alexandrov said currently available ‘point of care’ diagnostic tests which provided immediate results, such as blood glucose, pregnancy, and COVID test kits, used protein-sensing systems to detect the presence of sugar, pregnancy hormones, and COVID proteins.
“These, however, represent only a tiny fraction of what is needed in patient-focused health care model,” Professor Alexandrov said.
“However, developing new sensing systems is a challenging and time-consuming trial-and-error process.”
“The new ‘protein nano-switch’ method can massively accelerate development of similar diagnostics by decreasing the time and increasing the success rate. It uses proteins modified to behave like ON/OFF switches in response to specific targets.”
“The advantage of our approach is that the system is modular, similar to building with Lego bricks, so you can replace parts easily to target something else—another drug or a medical biomarker, for example.”
Professor Alexandrov said the method offered the possibility of building many different diagnostic and analytic tests, with a wide range of possible applications including diagnostics in human and animal health, testing kits for water contamination, and detecting rare earth metals in samples to direct mining efforts.
The multidisciplinary research team included scientists from QUT and the ARC Centre of Excellence in Synthetic Biology, consisting of lead researcher Professor Kirill Alexandrov, Dr. Zhong Guo, Cagla Ergun Ayva, Patricia Walden and Adjunct Professor Claudia Vickers.
The QUT team collaborated with leading electrochemists Evgeny Katz and Oleh Smutok from Clarkson University, in New York, and chemical pathologist Dr. Jacobus Ungerer from Queensland Health.
To demonstrate the technology, the team focused on a cancer chemotherapy drug that is toxic and requires constant measurement to ensure patient welfare.
“Too little of the drug won’t kill the cancer, but too much could kill the patient,” Professor Alexandrov said.
The sensor the team designed for the drug uses a color change to identify and quantify the drug.
Professor Alexandrov said the next step was the for the sensor to be tested in Queensland Health laboratories for approval for use in clinical setting.
“It’s really exciting, because it’s the first time an artificially designed protein biosensor may be actually suitable for a real-life diagnostic application” Professor Alexandrov said.
Dr. Ungerer said the protein-engineering technology developed by the research team provided a novel means to create laboratory tests.
“This has the potential to improve and expand laboratory testing, which will result in substantial health and economic benefits,” Dr. Ungerer said.
Dr. Guo said these advancements were made possible by an international and interdisciplinary team and excellent teamwork.
Professor Alexandrov said that the next step was to take this approach and standardize and scale it, to then start building more sophisticated sub-systems. He said there are two future directions for the work.
“One is to develop computer models that allow us to design and build the switches even more rapidly and precisely,” he said.
“The other is to demonstrate the scale and potential of the technology by building many switches for different diagnostic applications.”
Professor Alexandrov said the team were currently modifying existing proteins, but in the future, they could use the same principles to develop components that did not exist and would be designed from scratch.
“The new technique provides scientists unprecedented control over construction of protein-based sensing systems,” he said.
The article ‘Development of epistatic YES and protein logic gates and their assembly into signalling cascades’ is published in Nature Nanotechnology.
More information:
Guo, Z. et al. Development of epistatic YES and AND protein logic gates and their assembly into signalling cascades, Nature Nanotechnology (2023). DOI: 10.1038/s41565-023-01450-y. www.nature.com/articles/s41565-023-01450-y
Journal information:
Nature Nanotechnology
Provided by
Queensland University of Technology